1. Overview
This project portfolio documents Zhou Zegang’s contributions to project PatientBook. [PatientBook]
PatientBook is an all-in-one convenient desktop application for medical professionals in Singapore.
It is a Command Line Interface(CLI) application in which doctors can type simple commands on keyboard to store and
manage patients’ data and appointments’ details easily. It can also be used to retrieve information about
diseases, symptoms and drugs.
The T12-2 development team of PatientBook consists of five year-two computer science undergraduates from School of Computing,
National University of Singapore. [Team Info]
PatientBook supports various important operations such as finding information about a patient,
adding an appointment for a patient and predicting a disease from a set of symptoms.
2. Summary of Contributions
This section provides an overview of Zhou Zegang’s contributions to project PatientBook.
-
Code contributed: Code From Dashboard [Code]
-
Major enhancement: added diagnosis model which supports add disease, find disease, list disease and predict disease commands
-
What it does: This feature allows medical professionals to
adda disease and its related symptoms,findsymptoms of an existing disease,listall stored diseases andpredicta disease from a given set of symptoms in PatientBook. -
Justification: This feature improves the product significantly because a doctor can check up a disease’s symptoms and ascertain the accuracy of the diagnosis using
predictcommand. Thus, this enhancement will reduce the likelihood of medical malpractice to a large extent. -
Highlight: This enhancement is totally another separate model from two existing models, namely
patient modelandappointment model. As a result, it required an in-depth analysis of both design and integration with the other two models. The implementation too was challenging as it required changes to existing project structure. -
Credit: The
diagnosis modeluses an existing disease-symptom knowledge database fromColumbia University. [Database]
-
-
Other contributions:
-
Minor enhancement:
-
designed and added a generic method which converts a list of items to an
alphabetically orderedstring message so that it can be clearly displayed in the application. -
Resolved CSV file reading issue for drug utility function in jar . (Pull request: [#184])
-
-
Project management:
-
Documentation:
-
Community:
-
3. Contributions to the User Guide
Given below are sections I contributed to the User Guide. They showcase my ability to write documentation targeting end-users. |
Add a disease : add disease
Format: add disease d/DISEASE s/SYMPTOM
Adds a new disease to the patient book.
You must include one and only one DISEASE parameter with prefix d/ and at least one SYMPTOM for that disease
with prefix s/. Duplicate symptoms can be included but only one will be added into the patient book. The DISEASE
and SYMPTOM parameter can take any characters except comma. Both DISEASE and SYMPTOM parameters cannot exceed
20 characters in length.
When you try to add an existing disease into the patient book, the following error message will be shown to you:
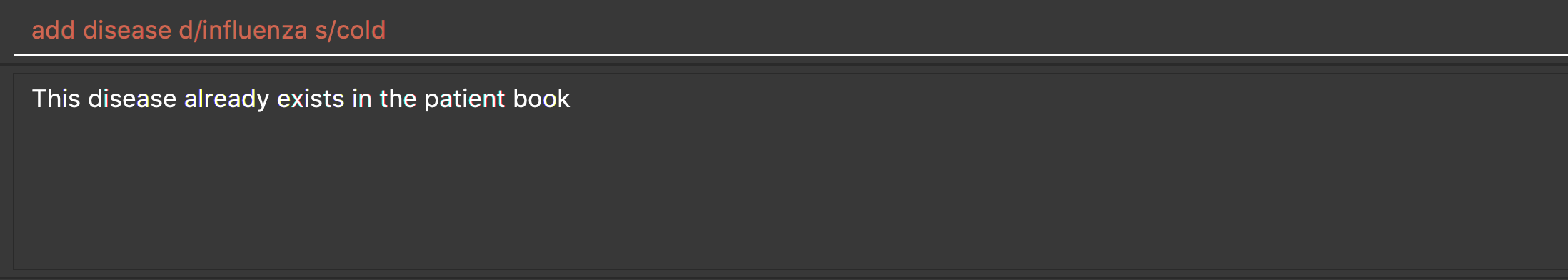
Hence, you may try command find disease NAME to find related symptoms of this existing disease.
Example:
-
add disease d/acne s/pustules s/blackheads

-
add disease d/alcohol misuse s/stroke s/stroke s/mouth cancer
Symptom 'stroke' will be included once in our record. -
add disease d/obesity d/obesities s/overweight
This is an invalid command due to multiple occurrence of disease paramters. Hence, you will be prompted to enter a correct command:
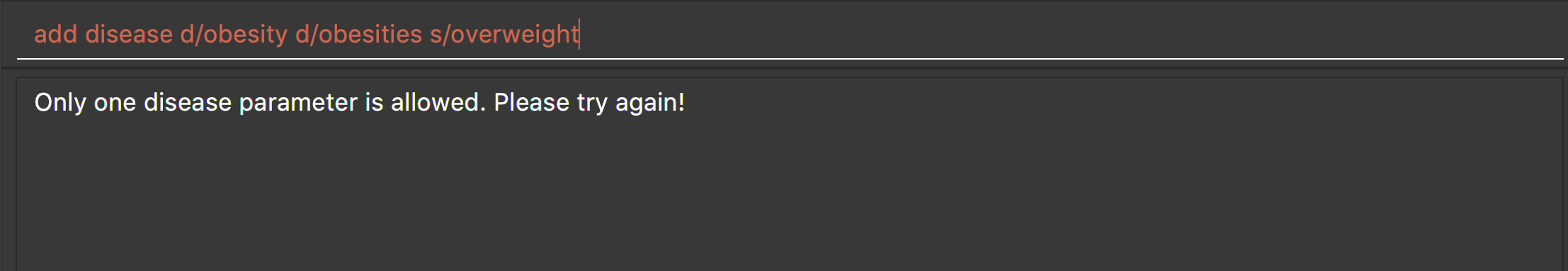
-
add disease d/averylongdiseaseparameter s/dummySymptom
Returns an error message:
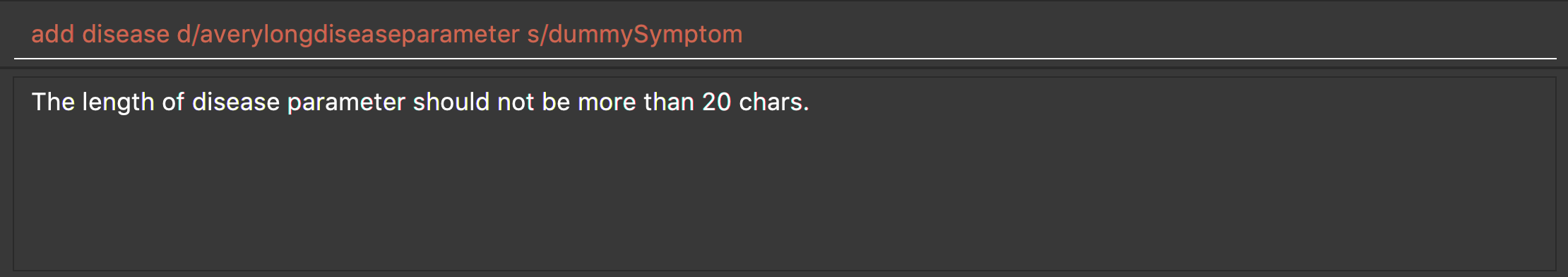
-
add disease d/invalid,disease s/dummySymptom
Returns an error message:
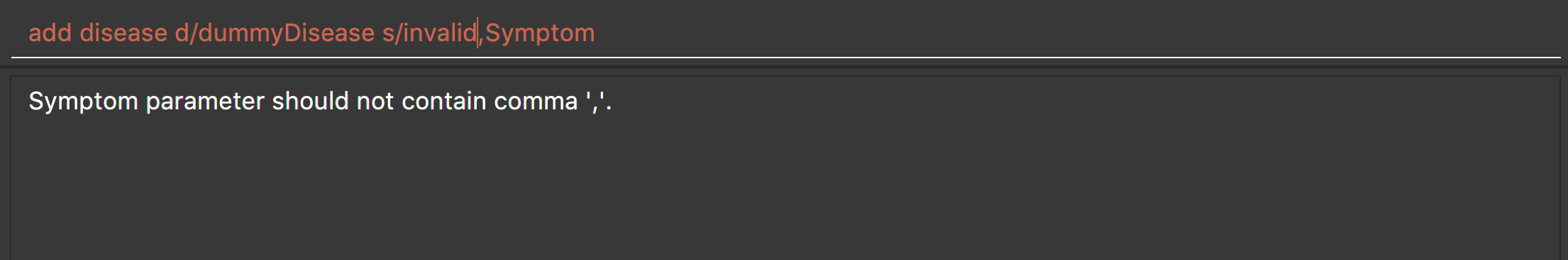
List all diseases : list disease
Format: list disease
Shows a list of all diseases in alphabetic order in the patient book.
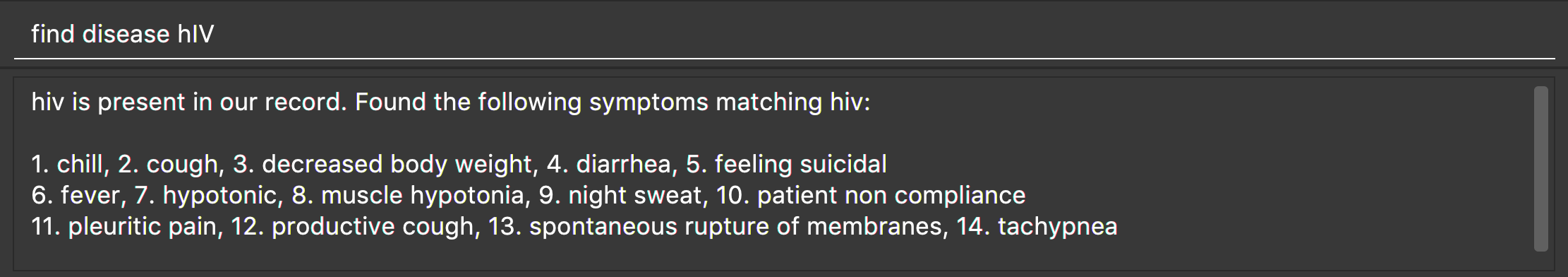
Locate diseases : find disease
Format: find disease NAME
Finds the disease which its name is specified in the command and returns its related symptoms displayed in alphabetic order.
-
The search is case insensitive; e.g. ‘HIV’, ‘hIV’ and ‘hiv’ are all equivalent.
-
Only full words will be matched; e.g. ‘confus’ will not match ‘confusion’.
Example:
-
find disease influenza
Returns all the symptoms related to disease ‘influenza’:

Predict disease : predict
Format: predict s/SYMPTOM
Predicts a disease from a given set of symptoms.
You must include at least one SYMPTOM parameter with prefix s/ to predict a disease. All
diseases that contain the set of symptoms will be returned and displayed in alphabetic order.
-
The
SYMPTOMparameter is case insensitive; e.g. ‘fever’ will match ‘FeVER’. -
Only diseases containing all the symptoms given in the command will be returned; e.g. For command
predict s/fever s/blackout, even if disease ‘bacteremia’ contains symptom ‘fever’, it will not be returned since it does not contain symptom ‘blackout’. -
If none of diseases contain the given set of symptoms, an error message will be shown to you:
Example:
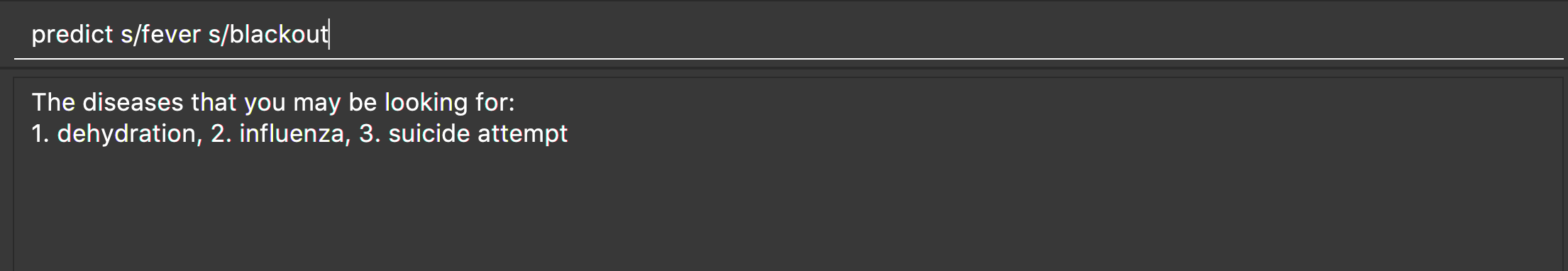
-
predict s/fever s/blackout
Returns diseases which contain the two symptoms:
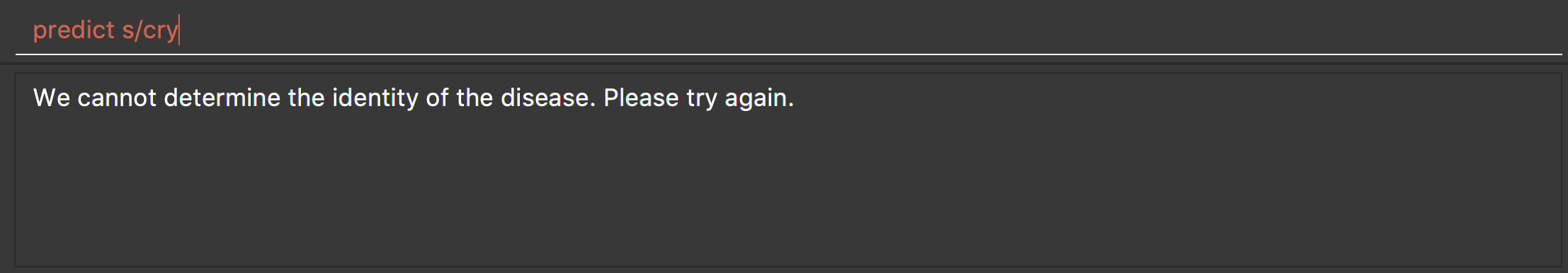
-
predict s/cry
Returns an error message:
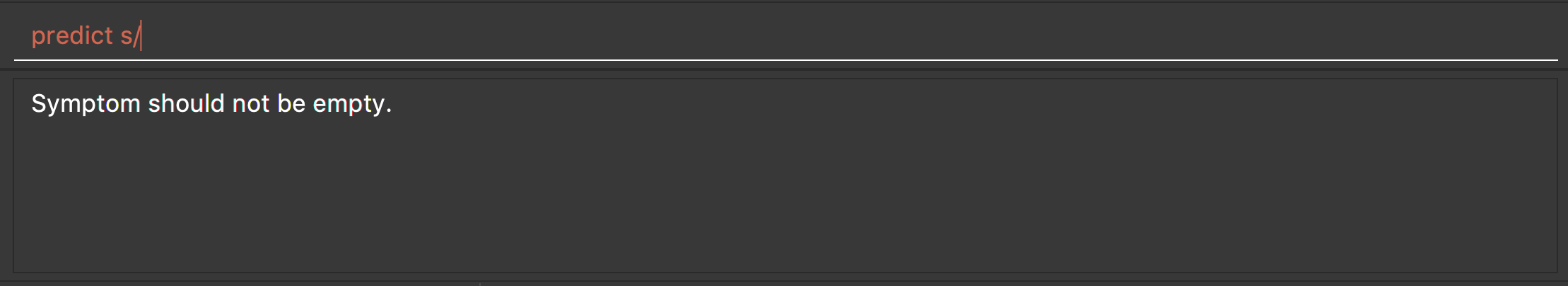
-
predict s/
Returns an error message:
4. Contributions to the Developer Guide
Given below are sections I contributed to the Developer Guide. They showcase my ability to write technical documentation and the technical depth of my contributions to the project. |
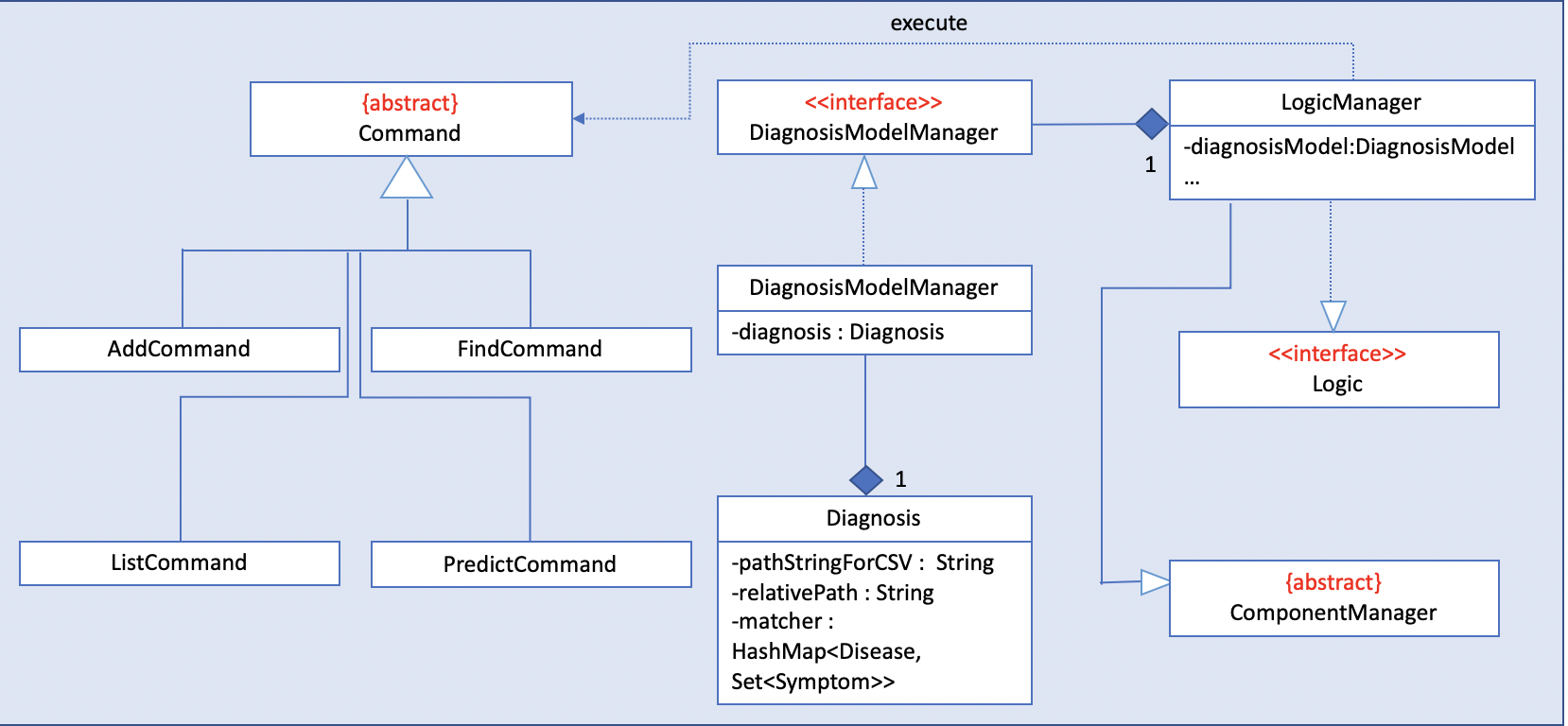
Diagnosis
Current Implementation
The diagnosis feature is facilitated by Diagnosis class. This class contains a private attribute called
matcher of type HashMap<Disease, Set<Symptom>>. Additionally, it implements the following operations:
-
Diagnosis#hasDisease(Disease disease)– Check if the application contains thediseaseinput by users -
Diagnosis#getSymptoms(Disease disease)– Return alistof all the relatedsymptomsof adiseaseinput by users -
Diagnosis#getDiseases()– Return alistof existingdiseasesfrom the database -
Diagnosis#addMatcher(Disease disease, Set<Symptom> symptoms)– Store a particulardiseasewith its set ofsymptomsin database. -
Diagnosis#predictDisease(Set<Symptom> symptoms)– Predict adiseasefor a set ofsymptomsinput by users.
These operations are exposed in the DiagnosisModel interface as
DiagnosisModel# hasDisease(Disease disease),
DiagnosisModel#getSymptoms(Disease disease), DiagnosisModel#getDiseases(),
DiagnosisModel#addMatcher(Disease disease, Set<Symptom> symptoms) and
DiagnosisModel#predictDisease(Set<Symptom> symptoms) respectively.
Given below is an example usage scenario and how the diagnosis mechanism behaves at each step:
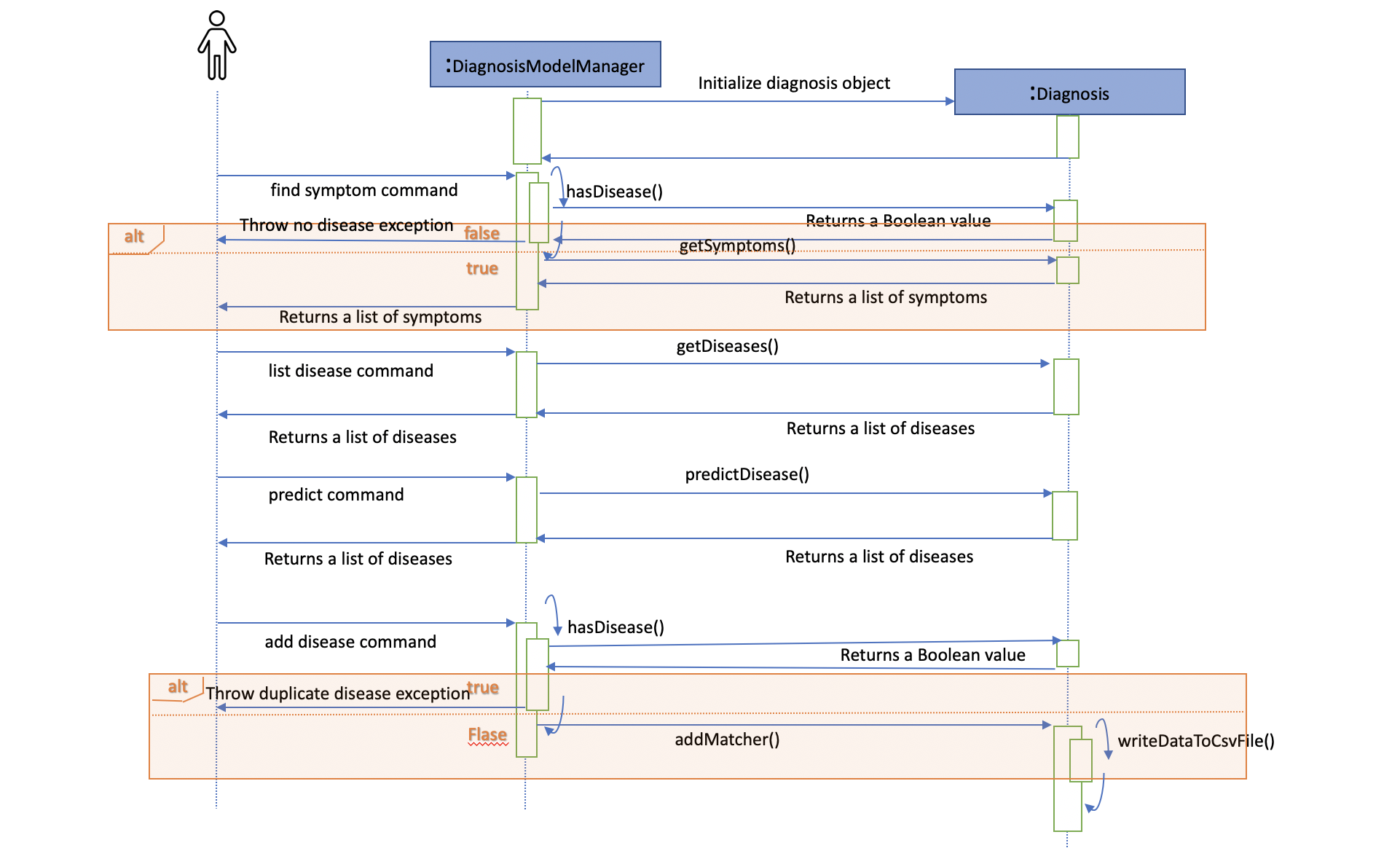
Step 1. The user launches the application for the first time. The Diagnosis will be instantiated and its
private attribute matcher will be initialized by calling the static method Diagnosis#readDataFromCsvFile().
Step 2. The user executes find disease Influenza command to get symptoms of “Influenza” stored in the patient book.
The find disease command calls DiagnosisModel#hasDisease(Disease disease) first, if the return Boolean value is false,
the user will be notified with a command exception thrown . If the return value is true, it will continue to call
DiagnosisModel#getSymptoms(Disease disease) and get all the symptoms related to “Influenza” in a List<Symptom>.
Step 3. If the disease is not present in the database record, the user can execute
add disease d/Influenza s/ncoordination s/fever s/pleuritic pain… command to insert the data into the database record.
Now, the add disease command calls DiagnosisModel#addMatcher(Disease disease, Set<Symptom> symptoms).
Now it will call the static method writeDataFromCsvFile(Disease disease, Set<Symptom> symptoms) which returns
a Hashmap of type HashMap<Disease, Set<Symptom>> and assigns it to this.matcher to update the private attribute.
/**
* Adds a disease and its related symptoms into the database
*
* @param disease disease input.
* @param symptoms related symptoms.
*/
public void addMatcher(Disease disease, Set<Symptom> symptoms) {
requireNonNull(disease);
requireAllNonNull(symptoms);
boolean hasDisease = this.hasDisease(disease);
if (!hasDisease) {
this.matcher = writeDataToCsvFile(disease, symptoms);
}
}Step 4. The user executes list disease command to get a list of diseases stored in the patient book.
The list disease command calls DiagnosisModel#getDiseases() which returns a list of diseases present in the
key set of matcher.
Step 5. Now the user decides to execute the command predict to search for a disease in database that
matches with input symptoms. This command calls DiasnosisModel#predictDisease(Set<Symptom>) which returns a list
of diseases that are mapped to a set of symptoms containing the given set.
/**
* Predicts a disease with a given set of symptoms.
*
* @param symptoms symptoms input.
* @return a list of qualified diseases.
*/
public List<Disease> predictDisease(Set<Symptom> symptoms) {
requireAllNonNull(symptoms);
List<Disease> diseases = this.matcher.keySet().stream()
.filter(disease -> this.matcher.get(disease).containsAll(symptoms)).collect(Collectors.toList());
diseases.sort(Comparator.comparing(Disease::toString));
return diseases;
}Design Considerations
Aspect: Disease-symptom matching data structures
-
Alternative 1 (current choice): Use a
HashMapwhere thekeyandvalueare the disease and set of related symptoms respectively.-
Pros: Better performance in terms of efficiency; duplicate values are also eliminated.
-
Cons: Difficult for other developers to understand.
-
-
Alternative 2: Define a
Matchclass with two private attributes representing the disease and a set of related symptoms, then maintain alistofMatchobjects.-
Pros: Easy for new developers to understand when disease-symptom pairs are encapsulated in an object.
-
Cons: May take up more memory, leading to performance issues.
-
Aspect: Searching algorithm to support disease prediction operation
-
Alternative 1 (current choice): Use
streamto search and collect all potential diseases.-
Pros: Easy to implement and easy for new developers to understand.
-
Cons: May have performance issues in terms of lower efficiency.
-
-
Alternative 2: Use a more systematic way to determine the suitability of each disease by calculating the similarity of its pre-existing set of symptoms and the set of symptoms given by users.
-
Pros: More accurate in determining the correct diseases.
-
Cons: Hard to implement and understand.
-